
Constantinos Chronis, PhD
Assistant Professor
Department of Biochemistry and Molecular Genetics
Pronouns: He/him
Contact
Building & Room:
MBRB 2370
Address:
900 S. Ashland Avenue, Chicago, IL 60607
Office Phone:
Email:
Related Sites:
Research Interests Heading link
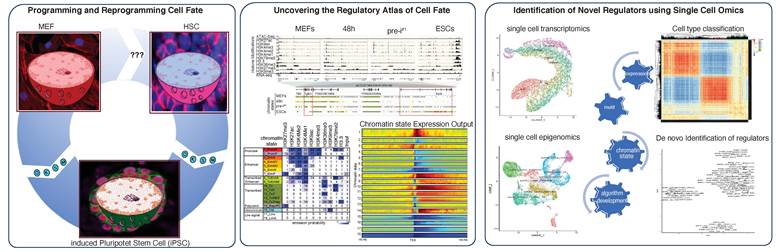
Cell fate specification is among the most fundamental processes in biology and is exerted physiologically during stem cell differentiation, pathologically during cancer stem cell formation, and can be engineered with reprogramming approaches.
All these processes involve activation of target programs and inhibition of donor cell programs and are controlled by sequence-specific DNA binding transcription factors (TFs) and their diverse interactions with the chromatin landscape.
We aim to understand how TFs decode mammalian genomes to generate the diverse cell identities that arise during development, cellular reprogramming and pathological conditions. To this end we employ multi-omic approaches, genome engineering, single cell tool development and bioinformatic analysis to delineate the TF-driven mechanisms that determine cell fate specification. Our ultimate goal is to engineer customized stem cells and their differentiated progeny for clinical applications while exploring the mechanisms that govern organ aging and regeneration and tumor growth.
Current Interests include
- A roadmap of mammalian development
How are pluripotent stem cells “programmed” to generate all bodily cell types during development, and how can somatic cells be “reprogrammed” to an ESC-like state?
We develop and apply innovative genomic technologies to map the epigenetic and transcriptomic events that drive cell fate decisions during early ESC differentiation and somatic to iPS reprogramming. - Reprogramming transcription factors and gene regulation
How can transcription factors identify and select cell type-specific genomic targets from thousands of potential binding sites?
Current research in the lab is aimed at understanding how reprogramming transcription factors facilitate the loss or destabilization of somatic cell identities and the activation of new target programs at single cell resolution - Engineering customizable cell fates
How can we engineer clinically relevant cell types to alleviate pathological conditions?
We use computational and systems biology approaches to define novel reprogramming TF combinations that can contribute to the generation of Hematopoietic Stem, Progenitor and Mature blood cell types with the ultimate goal of transplantation therapy and to understand mechanisms that contribute to blood diseases.
Selected Grants Heading link
- Norn Group -Impetus Grant Funding Mechanism. Defining a chromatin-state clock to measure and reverse hematopoietic stem cell aging – PI:Chronis
- NHLBI –R01HL170286 Reprogramming Gene Regulatory Networks to a Hematopoietic Stem Cell State – PI:Chronis
- NHLBI –P01 HL160469 The lung endothelium as an instructive niche for the innate immune system during vascular injury – Epigenetics and Transcriptomics Core Lead: Chronis
- High-Resolution Ensemble 3DStructures of Genome across Tissues INCITE award – Argonne National Labs – Co-PI: Chronis
Selected Publications
Dror I, Chitashvili T, Tan S.Y.X., Cano C.T., Sahakyan A, Markaki Y, Chronis C, Collier A, Deng W, Liang G, Sun Y, Afasizheva A, Miller J, Xiao W, Black D.L, Ding F, and Plath K. XIST directly regulates X-linked and autosomal genes in naïve human pluripotent cells. Cell 2023 In press.
Rho H, Terry AR, Chronis C & Hay N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metabolism 2023 35 (8), 1406-1423
Leung EHW, Joves K, Petenkaya A, Barham G, Henderson TG, Liang J, and Chronis C. Reprogramming cell fates towards novel cancer immunotherapies. Current Opinion in Pharmacology 2022 67,102312
Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, Chau A, Sabri S, Leonhardt H, Zheng S, Guttman M, Black DL, Plath K. A protein assembly mediates Xist localization and gene silencing. Nature. 2020 Nov;587(7832):145-151.
Zhang W*, Chronis C*, Chen X, Zhang H, Spalinskas R, Pardo M, Chen L, Wu G, Zhu Z, Yu Y, Yu L, Choudhary J, Nichols J, Parast MM, Greber B, Sahlén P, Plath K. The BAF and PRC2 Complex Subunits Dpf2 and Eed Antagonistically Converge on Tbx3 to Control ESC Differentiation. Cell Stem Cell. 2019 Jan 3;24(1):138-152.e8
Pasque V*, Karnik R*, Chronis C*, Petrella P, Langerman J, Bonora G, Song J, Vanheer L, Sadhu Dimashkie A, Meissner A, Plath K. X Chromosome Dosage Influences DNA Methylation Dynamics during Reprogramming to Mouse iPSCs. Stem Cell Reports. 2018 May 8;10(5):1537-1550.
Chronis C*, Fiziev P*, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. Cooperative Binding of Transcription Factors Orchestrates Reprogramming. Cell. 2017 Jan 26;168(3):442-459.e20
Denholtz M*, Bonora G*, Chronis C, Splinter E, de Laat W, Ernst J, Pellegrini M, Plath K. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013 Nov 7;13(5):602-16
Sridharan R, Gonzales-Cope M*, Chronis C*, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M, Carey M, Garcia BA, Plath K. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat Cell Biol. 2013 Jul;15(7):872-82
Education
• BSc(Hons),University of Edinburgh, UK
• MSc by Research (Distinction), University of Edinburgh, UK
• PhD, Kings College London, UK
• Research Fellow: University of California, Los Angeles, CA (Plath lab)
Professional Memberships
ISSCR, ASH member